Hi. We would like to functionally annotate some metagenomic sequencing reads without doing any assembly or binning or read mapping to reference genomes. Just annotate by searching things like nr, PFAM, COGs, etc. Anyone know of tools that do this?
— Jonathan Eisen (@phylogenomics) March 29, 2023
Humann suggestion
fraggenescan on the raw reads, then use hmmer w/ pfam-A or kegg hmm.
EggNog Mapper
MGRAST
Metawrap
Yooseph approach
https://pubmed.ncbi.nlm.nih.gov/27400380/
https://pubmed.ncbi.nlm.nih.gov/27585568/
mi-faser
https://bromberglab.org/project/mifaser/…
MGX
https://microbiomejournal.biomedcentral.com/articles/10.1186/s40168-018-0460-1…
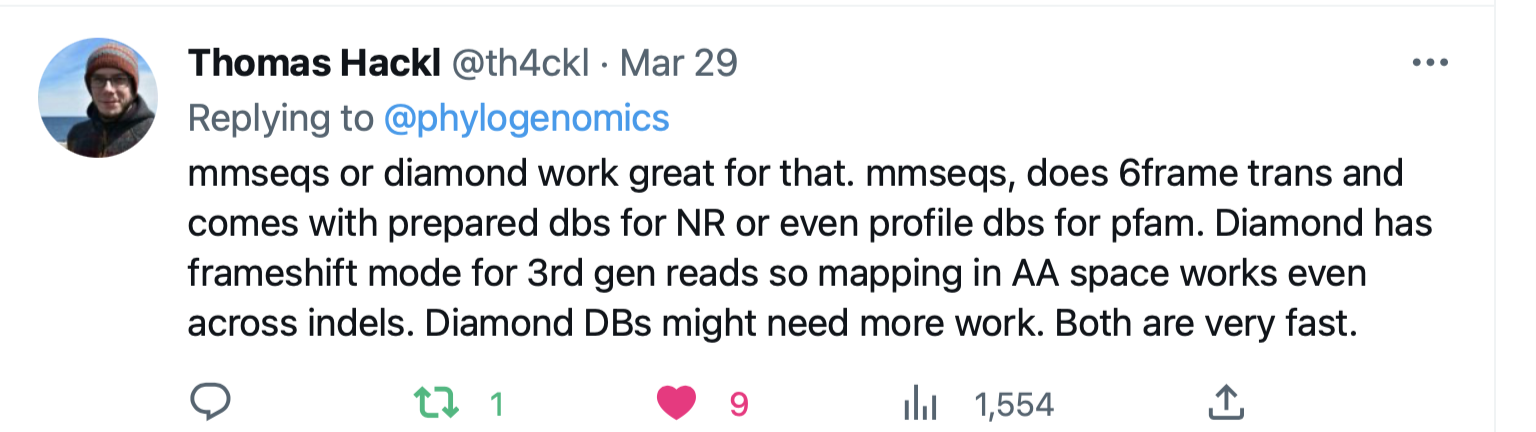
MMseqs or Diamond
Squeeze Meta
https://github.com/jtamames/SqueezeMeta
Metageni/Superfocus
https://github.com/metageni/SUPER-FOCUS
Plass
https://github.com/metageni/SUPER-FOCUS
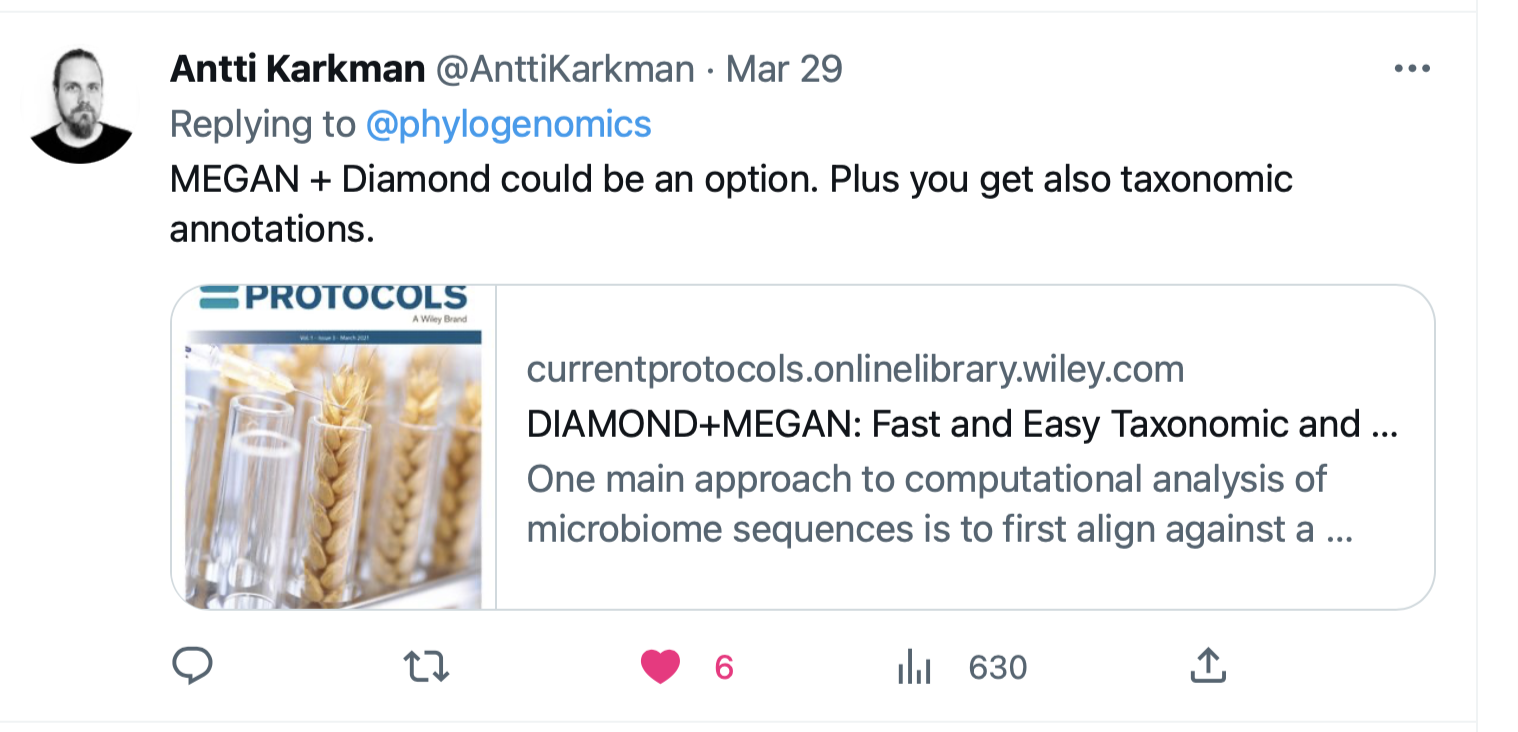
Megan + Diamond
https://currentprotocols.onlinelibrary.wiley.com/doi/full/10.1002/cpz1.59
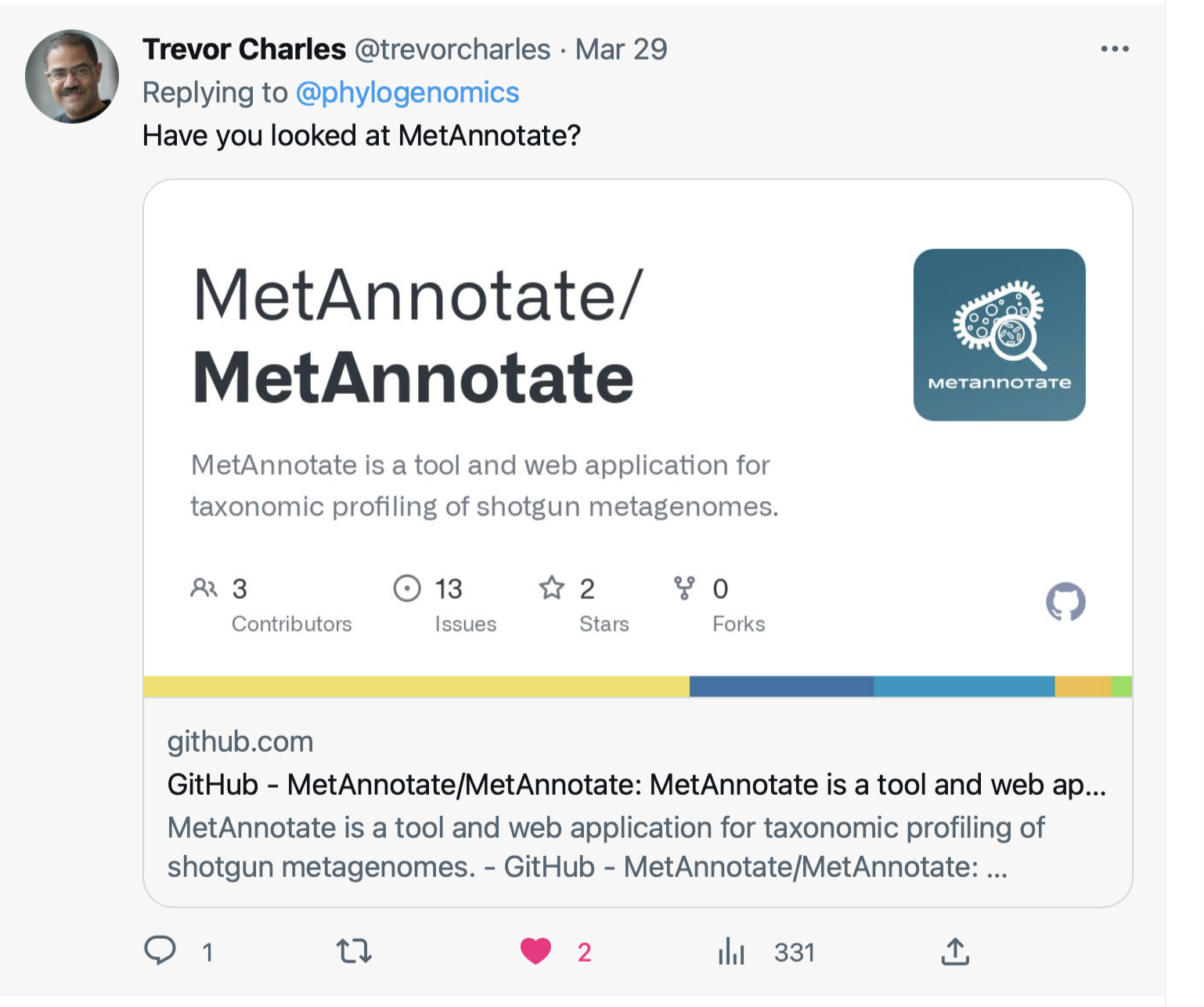
MetAnnotate
https://github.com/MetAnnotate/MetAnnotate
Review of relevance
https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-019-6289-6
UPROC
https://pubmed.ncbi.nlm.nih.gov/25540185/
Looking Glass
https://www.nature.com/articles/s41467-022-30070-8
Shortbred
Proteinfer / MGnify
https://www.ebi.ac.uk/metagenomics/pipelines/2.0
AntiSmash
MMseq2